CircRNA as an emerging role in acute myocardial infarction: a literature review
Introduction
Background
In recent years, the incidence, disability and mortality of cardiovascular disease (CVD) have increased dramatically, and acute myocardial infarction (AMI) is the main cause of death of CVD (1,2). AMI is the most common disease in CVD, with the highest fatality rate. About 85% of deaths in CVD are caused by AMI and stroke (3,4). In the past, it was generally believed that AMI mainly occurred in middle-aged and elderly people, but with the development of society and the change of lifestyle, the incidence of AMI gradually showed a trend of younger and increased year by year (5). In most countries around the world, AMI has become the main role of chronic diseases and the source of huge economic burden in society (6,7). AMI is a multifactorial disease involving complex pathophysiological processes, which is caused by partial or complete coronary artery occlusion and results in ischemic injury of cardiomyocytes in the corresponding areas of the heart (8). Once myocardial cells are damaged by ischemia, the heart will be further fibrosis and remodeling, and eventually develop into chronic heart failure (CHF) or even sudden cardiac death (SCD) (9-11). With the development of medical technology, great progress has been made in the diagnosis and treatment of AMI. Although reperfusion therapy is the most effective treatment at present, which can reduce the mortality of patients with AMI, many survivors will still suffer from myocardial remodeling and heart failure (HF) (12). Yin et al. believe that effective therapeutic targets of protect cardiomyocytes from apoptosis and fibrosis are still limited, and that it is necessary to further understand of the pathogenesis of AMI at the molecular level, in treatment of AMI (2). In the past decades, many molecules and signaling pathways have been identified as potential therapeutic targets. however, a new therapeutic strategy of promote cardiac repair in patients with AMI have yet to be realized (13,14). Therefore, there is still a long way to go to explore the pathogenesis of AMI and seek a more perfect treatment strategy.
Rationale and knowledge gap
CircRNA was discovered and reported in 1976. As a non-classical RNA splicing product, circRNA was once considered as a by-product by incorrect splicing (15). CircRNA is a kind of non-coding RNA, which contains a closed continuous loop structure and is widely expressed in the genomes of multiple species (16). CircRNA is highly evolutionarily conservative at the sequence level between different species, and widely exists in heart tissue (17). In recent years, based on advanced RNA sequencing technology and bioinformatics analysis technology, circRNA has become a hot and frontier area in RNA research. However, their functions in cells have only been newly recognized recently (18). At present, circRNAs have been widely studied as biomarkers for cancer, such as gastric cancer (GC), hepatocellular carcinoma (HCC) and lung cancer (19). Some circRNAs may also become biomarker candidates for age-related diseases, such as CVD, neurodegenerative diseases, Alzheimer’s disease, aging, etc. (19,20). Many studies have confirmed that the abnormally expressed circRNA is closely related to the occurrence and development of CVD, such as AMI, myocardial fibrosis (MF), cardiac aging, myocardial hypertrophy, HF and coronary atherosclerosis (21-26). Studies have also shown that circRNA is involved in diseases other than AMI, such as atherosclerosis, essential hypertension, chronic thromboembolic pulmonary hypertension, dilated cardiomyopathy and hypertrophic cardiomyopathy, congenital heart disease in children, aortic aneurysm and thoracic aortic dissection, and retinal vascular dysfunction caused by diabetes (24-30). There was a bioinformatics study have detected circRNA in three patients with AMI and three patients in control group through gene chip. This study found that there are 650 circRNAs differentially expressed in AMI, and that circRNA-miRNA interaction pathway is very likely to participate in regulating the occurrence and development of AMI (2). However, the relationship between circRNAs and AMI, and its clinical value have not yet been fully clarified. Therefore, we summarized the application value of circRNA in AMI.
Objective
Our review aims to: (I) summarize the biological characteristics and involvement in the pathophysiology of circRNA in AMI; (II) discuss that the circRNA may become a biomarker and therapeutic target for AMI in the future; (III) provide potential clinical insights into new strategies for the diagnosis and treatment of AMI; (IV) and provide prospects for further research on circRNA in AMI. We present this article in accordance with the Narrative Review reporting checklist (available at https://jxym.amegroups.com/article/view/10.21037/jxym-23-10/rc).
Methods
We searched the PubMed database for original studies and review articles published from January 1993 to June 2022. We carefully read, analyzed and summarized related research literature and some key citations. The terms were circRNA, Circular RNA, AMI, acute myocardial infarction, biomarkers, myocardial apoptosis, and pathophysiology, all of which were used in different combinations. The retrieval strategy is summarized in Table 1 and Table S1.
Table 1
| Items | Specification |
|---|---|
| Date of search | From August 14 to November 12, 2022 |
| Databases and other sources searched | PubMed |
| Search terms used | Detailed search terms are shown in Table S1 |
| Timeframe | From January 1993 to June 2022 |
| Inclusion and exclusion criteria | Inclusion criteria: original and critical articles and some of their key citations s in English languages |
| Exclusion criteria: (I) studies that duplicate reports; (II) studies with incomplete data; and (III) case reports, summaries of meetings and letters | |
| Selection process | Two authors evaluated and selected the studies independently. If there is any disagreement, it will be resolved by all authors through consultation and discussion |
The biological characteristics of circRNA
The biogenesis of circRNA
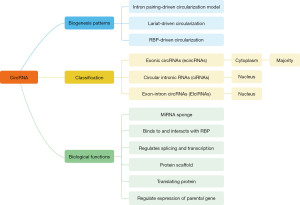
CircRNA is a naturally formed endogenous non-coding RNA. Unlike traditional linear RNA, circRNA presents a covalent closed-loop structure. It is not easy to be degraded by exonuclease and has high abundance, high stability and specificity at the stage of tissue development, and conservation of evolution between species, because it does not have a 5' cap end and a 3' ploy (A) tail end (31). CircRNA is widespread in cells and tissues. The expression of some circRNAs is more than ten times higher than that of typical linear transcripts generated by the same genes (17). CircRNA usually exhibits specific expression pattern at the stage of cell and tissue development (32,33), suggesting that they may be involved in some physiological and pathological processes. Some circRNAs also have internal ribosome entry sites (IRESs), which provide potentiality for protein synthesis (34,35). There are mainly two classical biogenesis models: lasso driven cyclocyclic and intron paired driven cyclocyclic (17). An alternative biogenesis model has also been proposed, which formed a bridge between flanking introns by binding with RNA-binding protein (RBP) to make splicing donors and receptors close, thus promoting the cyclization of exons (36-38). Its biogenetic characteristics are summarized in Figure 1. These characteristics indicate that circRNAs are potential regulators of transcription and post-transcription (19). In conclusion, circRNA may play a specific role in the occurrence and development of some diseases.
The biological expression of circRNA
RNA sequencing of human adult and fetal tissues (heart, kidney, liver, lung, colon and stomach) showed that up to 50% of circRNA had tissue specificity, and both the number and expression level of circRNA in fetal tissues were higher than those in adult tissues (24,39). CircRNA is the most abundant in the brain, compared with other organs, thus stress and aging play an important role in changing its expression profile (39-41). CircRNA is abundant and stable in body fluids such as peripheral serum or plasma, urine, saliva, cerebrospinal fluid, milk and circulating blood cells, tumor cells and exocrine bodies (20,42). Sequencing data revealed that there are more than 15,000 circRNAs in human heart (43). Tan et al. performed deep sequencing of RNA isolated from 14 human hearts and 25 mouse hearts, and 15,318 and 3,017 circRNAs were found respectively. The most abundant circRNA expressed in the heart is CircSLC8A1-1 located in the cytoplasm (43). Stanislas Werfel et al. sequenced circRNAs in human, mouse and rat hearts, which shows that circRNAs are enriched and differentially expressed in postnatal development and heart diseases (44).
The classification of circRNA
As summarized in Figure 1, circRNAs can be divided into three categories based on their biogenesis and sequence: exonic circRNAs (ecircRNAs), circular intronic circRNAs (ciRNAs), and exon-intron circRNAs (EIciRNAs). (I) The ecircRNAs are derived from exons in linear transcripts and lack any introns in their sequences. Most circRNAs belong to this group and mainly exist in the cytoplasm. (II) The ciRNA is present in the nucleus, which is not generated by backsplicing and lacks exon sequence and has little enrichment for miRNA target sites. (III) The EIciRNA contains both introns and exons in its sequence, which is mainly localized to the nucleus and interacts with U1snRNP and Pol II, thereby promoting the transcription of its parental genes (17,19,45-48). It has also been reported that circRNAs can also be divided into intragenic and intergenic according to the position in the genome. Interestingly, ecircRNA, ciRNA and ElciRNAa are all intragenic circRNAs, which are derived from the sequences in the parental loci (49,50).
The biological function of circRNA
According to previous studies, circRNAs regulate gene expression mainly by binding to miRNAs and causing miRNA dysfunction. As shown in Figure 1, circRNAs act as miRNA sponge, which can competitively bind miRNAs and inhibit the regulation of miRNAs on their target gene mRNA, thereby indirectly inhibiting or promoting the expression of mRNA. Binding to RBP acts as target RNA bait, as a modulator of transcription and splicing by binding srnRNA and upgrading Pol II activity, and as a modifier of protein scaffold and parental gene expression (18,32,36,45). In the past, it was generally believed that circRNA act as a non-coding RNA could not translate proteins. But in recent years, a few studies have reported that circRNA molecules can be translated into proteins or peptides when they contain IRES elements or prokaryotic ribosome binding sites (45,51-53). For example, human Circ-ZNF609 is derived from the second exon of its host ZNF gene, which is translated into a protein in a splicing-dependent and cap-independent manner (34,54). Legnini et al. confirmed that Circ-ZNF609 is associated with heavy polyribosomes and provided an example of circRNA encoding protein in eukaryotes (34,53).
CircRNA in AMI
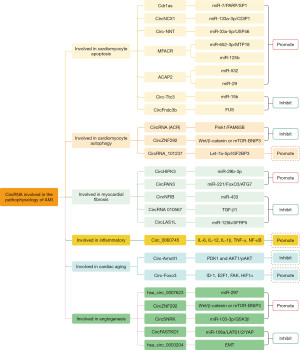
Apoptosis and necrosis of cardiomyocyte after AMI induce inflammatory response, MF, myocardial remodeling gradually occur, and finally develop into HF, while neovascularization promotes tissue repair after MI. Therefore, it is possible to explore a new approache to the treatment of AMI if our research focuses on cell apoptosis, MF, regulation of inflammatory response, and angiogenesis after AMI. Many circRNA have been found to be involved in regulating the above pathophysiological processes after MI (55-59) (as summarized in Figure 2).
As shown in Table 2, some circRNAs were differentially expressed in AMI, which involved the pathophysiological process of AMI. Overexpression or knock out some circRNAs may aggravate or alleviate AMI. As shown in Table 3, some circRNA have also been identified as biomarker of AMI. This will provide new insights into the diagnosis and treatment strategies for AMI. However, most of these studies were conducted on animals, using rats or mice as specimens.
Table 2
| Influence | CircRNA | Species | Samples | Expression | Pathway | Function | Ref. |
|---|---|---|---|---|---|---|---|
| Aggravating AMI | Cdr1as | Mice | Cardiomyocyte | Up | Cdr1as-miR-7a-PARP/SP1 | Promote cell apoptosis, increase MI size | (60) |
| MFACR | Mice | Cardiomyocyte | Up | miR-125b-MTP18 and miR-652-3p | Promote cardiomyocyte apoptosis and MI | (61,62) | |
| ACAP2 | Human/ rats |
Plasma/cardiomyocyte | Up | miR-532/miR-29 | Promote cardiomyocyte apoptosis | (63,64) | |
| Circ-NNT | Mice | Cardiomyocyte | Up | miR-33a-5p/USP46 | Promote myocardial apoptosis and I/R injury | (65) | |
| CircRNA_101237 | Mice | Cardiomyocyte | Up | let-7a-5p/IGF2BP3 | Promote apoptosis and autophagy | (66) | |
| CircHIPK3 | Mice | Cardiac fibroblasts | Up | miR-29b-3p | Promote myocardial fibrosis | (67) | |
| Circ_0060745 | Mice | Cardiomyocyte | Up | Inhibit NF-κB activity | Promote inflammation, increase MI area, deteriorate cardiac function | (68) | |
| Foxo3 | Mice | Cardiac tissue | Up | ID-1, E2F1, FAK, HIF1α | Induce senescence | (69) | |
| CircPAN3 | Rats | Cardiac fibroblasts | Up | miR-221/FoxO3/ATG7 | Promote myocardial fibrosis | (70) | |
| CircROBO2 | Mice | Cardiomyocyte | Up | miR-1184/TRADD | Promote cell apoptosis, increase MI size | (71) | |
| CircFASTKD1 | Human | Endothelial cells | Down | miR106a | Inhibiti angiogenesis | (72) | |
| hsa_circ_0003204 | Human | HUVEC | Up | Regulate of EMT | Slow the repair of endothelial cell injury | (73) | |
| Attenuating AMI | CircNcx1 | Mice | Cardiomyocyte | Up | miR-133a-3p/CDIP1 | Knockout: reduce cardiomyocyte apoptosis and I/R injury | (74) |
| Circ-Ttc3 | Rats | Cardiomyocyte | Up | miR-15b | Reduce cardiomyocyte apoptosis and cardiac dysfunction | (21) | |
| CircACR | Mice | Cardiomyocyte | Down | ACR-Pink1-FAM65B | Inhibit myocardial autophagy, protection I/R damage, reduce MI size | (75) | |
| CircRNA ZNF292 | Rats | Cardiomyocyte | Up | Wnt/β-catenin or mTOR-BNIP3 | Inhibiti apoptosis and autophagy | (76) | |
| CircNFIB | Mice | Cardiac fibroblasts | Down | miR-433 | Inhibiti myocardial fibrosis | (77) | |
| CircRNA 010567 | Rats | Cardiomyocyte | − | Inhibit TGF-β1 | Inhibiti apoptosis and myocardial fibrosis | (78) | |
| Circ_LAS1L | Human | Cardiac fibroblasts | Down | miR-125b/SFRP5 | Inhibiti myocardial fibrosis | (79) | |
| Circ-Amotl1 | Human | Cardiac tissue | Up | AKT1 phosphorylation, pATK nuclear translocation | Enhance cell proliferation and survival, protect Dox induced cardiomyopathy | (80) | |
| hsa_circ_0007623 | Human | Huvec | Up | miR-297 | Promotes cardiac repair | (81) | |
| CircSNRK | Rats | Cardiomyocyte | Down | miR-103-3p/GSK3 β/β-catenin | Inhibits apoptosis, induces angiogenesis, promotes cardiac repair | (82) | |
| CircFndc3b | Mice | Cardiac tissue | Down | FUS/VEGF-A | Reduces cardiomyocyte apoptosis, promotes angiogenesis, limits infarct size, improves cardiac function | (83) | |
| CircMACF1 | Mice | Cardiomyocyte | Down | miR-500b-5p/EMP1 | Repair cardiomyocyte apoptosis,reduce MI size | (84) | |
| CircPostn | Human/mice | Plasma/cardiomyocyte | Up | miR-96-5p/BNIP3 | Beneficial to H/R induced cardiomyocyte apoptosis, injury and remodeling | (85) | |
| CircNfix | Rats | Cardiomyocyte | Up | miR-214/Ybx1 | Deletion: induces myocardial regeneration and angiogenesis,inhibits cardiomyocyte apoptosis | (86,87) | |
| CircCDYL | Mice | Cardiomyocyte | Down | miR-4793-5p/APP | Promote cardiomyocyte proliferation, induce cardiac regeneration,improve prognosis | (88) |
AMI, acute myocardial infarction; MI, myocardial infarction; I/R, ischemia/reperfusion; HUVEC, Human Umbilical Vein Endothelial Cell; H/R, hypoxia/reoxygenation.
Table 3
| CircRNA | Species | Samples | Expression | Pathway | Function | Ref. |
|---|---|---|---|---|---|---|
| CircTMEM165, CircUBAC2, CircZNF609, CircANKRD12, CircSLC8A1 | Human | Blood | Up | – | Improve the accuracy of AMI diagnosis | (18) |
| CircRNA_081881 | Human | Plasma | Down | miR-548/PPARγ | Target for accurate diagnosis and treatment | (89) |
| hsa_circRNA_001654, hsa_circRNA_091761, hsa_circRNA_405624, hsa_circRNA_406698 | Human | Coronary blood | Up | has-miR-491-3p, has-miR-646, has-miR-603, has-miR922/RUNX1 | Biomarkers for the diagnosis of AMI | (90) |
| Circ-RCAN2 | Pig | Cardiac tissue | Down | – | Diagnostic value for AMI | (91) |
| CircSLC8A1, CircNFIX | Rats | Cardiomyocyte | Up | – | Diagnostic markers of SCD caused by IHD | (87) |
| MICRA | Human | Blood | Down | microRNA-150 | Predict LV dysfunction | (92,93) |
| CircNfix | Rats | Cardiomyocyte | Up | miR-214/Ybx1 | Diagnostic biomarker | (86,87) |
| CircPAN3 | Rats | Cardiac fibroblasts | Up | miR-221/FoxO3/ATG7 | Potential biomarker for cardiac fibrosis therapy | (70) |
| CircROBO2 | Mice | Cardiomyocyte | Up | miR-1184/TRADD | Provide evidence for clinical prognosis | (71) |
| CircFASTKD1 | Human | Endothelial cells | Down | miR106a | Biomarkers for AMI prognosis | (72) |
| hsa_circ_0124644 | Human | Peripheral blood | Up | – | Can be used as a diagnostic biomarker of CAD | (94) |
AMI, acute myocardial infarction; SCD, sudden cardiac death; IHD, ischemic heart disease; LV, left ventricle; CAD, coronary artery disease.
Involved in the pathophysiological process of AMI
Involved in cardiomyocyte apoptosis after MI
It has been found that the overexpressed Cdr1as in cardiomyocyte of mice after infarction can promote apoptosis of cardiomyocyte and increase the area of MI (26,60). Overexpression of CircFndc3b reduced the level of cardiomyocyte apoptosis in the MI mouse model (83). CircNCX1 is significantly upregulated in H2O2 treated H9c2 cells and ischemic myocardium, and knockdown CircNCX1 could reduce cardiomyocyte apoptosis to alleviate myocardial injury after ischemia/reperfusion (I/R) in rats (74). CircRNA (MFACR) associated with mitochondrial division and apoptosis regulates mitochondrial fission and apoptosis in the heart by directly down-regulating miR-652-3p (61). It has also been found that overexpressed MFACR increases cardiomyocyte apoptosis, and that MFACR may promote cardiomyocyte apoptosis and the progression of MI by down-regulating miR-125b (62). The overexpression of ACAP2 may increasing apoptosis after MI (63,64). The overexpression of Circ-Ttc3 protects cardiomyocytes from ischemia-related apoptosis, while silencing Circ-Ttc3 could aggravates cardiac dysfunction after AMI (21). Circ-NNT regulates USP46 by acting as miR-33a-5p sponge, thereby promoting apoptosis and cardiomyocyte I/R injury (65).
The above examples show that the great progression have been made in the study of apoptosis and circRNAs after AMI. We can hypothesize that silence or overexpress some certain circRNAs will reduce cardiomyocyte apoptosis after MI and improve prognosis.
Involved in cardiomyocyte autophagy after MI
Zhou et al. reported that autophagy related circRNA (ACR) inhibits cardiomyocyte autophagy and cell death by targeting PINK1 mediated FAM65B phosphorylation, and that ACR protects the heart from I/R injury and reduces the size of MI (75). CircZNF292 involved in angiogenesis and proliferation of endothelial cells and inhibits oxygen-glucose deprivation (OGD) induced apoptosis and autophagy (76,95). Knock out of CircRNA_101237 inhibited cardiomyocyte apoptosis through the autophagy pathway. CircRNA_101237 can regulate cardiomyocyte apoptosis and autophagy induced by anoxia/reoxygenation (A/R) injury (66).
Involved in MF after MI
Studies have confirmed that silence CircHIPK3 attenuated cardiac fibroblasts (CFs) proliferation and migration induced by AngII. The overexpressed CircHIPK3 can effectively reverse the inhibition of proliferation and migration of CFs by miR-29b-3p, and the combination of CircHIPK3 silence and miR-29b-3p overexpression has a stronger inhibitory effect on MF (67). Overexpressed CircNFIB decreased the proliferation of NIH/3T3 cell and CF, and inhibit CircNFIB promoted CFs proliferation. CircnFIB-miR-433 axis can be used as a new way to inhibit MF (77). In the AMI rat model, circRNA 010567 siRNA was found to improve cardiac function, reduce MF and inhibit cardiomyocyte apoptosis (78). Knockout of CircPAN3 reduced MF after MI. CircPAN3 shows a promoting effect in the process of MF, which can be used as a potential biomarker for MF treatment (70). CircLAS1L was downregulated in patients with AMI and CFs, and inhibits the activation, proliferation and migration of CFs. The CircLAS1L may play an important role in the process of MF after MI (79).
So in the same way, silencing or overexpressing of MF related circRNA may regulate cardiac function, promote cardiac repair and improve prognosis after MI.
Involved in inflammatory after MI
The ischemic injury of cardiomyocyte after AMI triggers a series of inflammatory responses, and the expression of certain circRNAs is regulated by oxidative stress. These circRNA mediate the generation of reactive oxygen species (ROS) and promote inflammation, cell death and apoptosis. For example, The expression of Circ_0060745 was significantly increased in the myocardium of mice with AMI. Knock out Circ_0060745 inhibited the apoptosis of cardiomyocytes and the expression of IL-6, IL-12, IL-1β, TNF-α and NF-κB. Overexpress Circ_0060745 resulted in increased infarct size and deteriorate cardiac function after AMI. Knock out Circ_0060745 reduced infarct size and improved cardiac function after MI by inhibiting cardiomyocyte apoptosis and inflammation (68).
Involved in angiogenesis after MI
Angiogenesis after AMI promotes cardiac repair, improves cardiac function and prognosis. Studies indicated that silence hsa_circ_0007623 could reduce cell proliferation, migration and angiogenesis in hypoxia induced human umbilical vein endothelial cells (HUVECs). hsa_circ_0007623 could promotes cardiac repair and protects cardiac function after acute myocardial ischemia (81). It has also been found that hsa_circ_0003204 may be involved in angiogenesis by regulating epithelial mesenchymal transition (EMT), and may delay the repair of endothelial cell injury (73). Boeckel et al. found that silence CircZNF292 reduced the tube formation and bulb sprouting of endothelial cells, and confirmed the role of CircZNF292 in promote angiogenesis (95). Overexpress CircSNRK could inhibit cardiomyocyte apoptosis, induce angiogenesis and cardiac regeneration, significantly restore myocardial function, and improve prognosis after MI (82).
In addition, Zeng et al. found that Circ-Amotl1 was highly expressed in the neonatal heart, increased cardiomyocyte survival, reduced apoptosis, and played a protective role in doxorubicin (Dox)-induced cardiomyopathy (80). Circ-Foxo3 was highly expressed in the hearts of elderly patients and mice, and overexpress CircFoxo3 could induce aging (69). At present, there are relatively few studies on circRNA and cardiac aging, which is a new field.
In summary, we can consider silencing or overexpressing some specific circRNAs to reduce myocardial injury, promote cardiac repair, improve cardiac function and prognosis from all aspects of apoptosis, inflammation, autophagy, fibrosis and angiogenesis. It may be a promising therapeutic target for the recovery of cardiac function after MI.
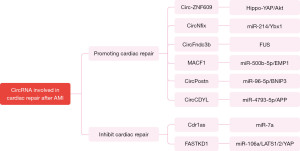
Involved in cardiac repair after AMI
At present, circRNA has been observed to promote or inhibit cardiac repair in many animal studies (summarized in Figure 3 and Table 2). Wang et al. revealed the role of Circ-ZNF609 in myocardial I/R injury. Knockdown of Circ-ZNF609 prevented acute I/R injury and attenuated LV dysfunction after I/R and contributes to cardiac repair (96). A study has identified that CircNfix overexpress in heart of human, rat and adult mouse. Down-regulation of CircNfix could promote cardiomyocyte proliferation and angiogenesis, inhibit cardiomyocyte apoptosis, and alleviate cardiac dysfunction after MI, ultimately promotes cardiac repair and functional recovery after AMI (86). In a previous study, circRNA microarray analysis was performed in mouse heart three days after AMI, which suggest that the overexpression of CircFndc3b could reduce cardiomyocyte and endothelial cell apoptosis, enhances neovascularization, limits infarct size, regulates cardiac repair after MI and improves myocardial function (83). Therefore, we can consider that knockdown of Circ-ZNF609 may serve as a promising therapeutic target to reduce myocardial I/R injury. CircNfix and CircFndc3b may be effective therapeutic target for restoring cardiac function after AMI.
The overexpressed Cdr1as promoted apoptosis of cardiomyocyte and increased infarct size in mice after MI (60). Interestingly, another study confirmed that the high tissue level of circRNA CDR1as in the AMI region of pig hearts was significantly correlated with left ventricular and right ventricular ejection fraction (LVEF, RVEF), and LV stroke output, and negatively correlated with infarction size (97). The overexpressed CircMACF1 could effectively repair cardiomyocyte apoptosis, significantly reduce infarct size and inhibit the progression of AMI (84). Down-regulation of CircFASTKD1 could promote angiogenesis, thereby improve the prognosis of MI (72). Knocking out CircPostn significantly reduced myocardial injury and infarct size in AMI mice, significantly increased LVEF, and inhibited the thickness of LV anterior and posterior wall diastolic (85). The overexpression and downregulation of CircCDYL after AMI improved and worsened the cardiac function, promoted and inhibited the proliferation of cardiomyocytes in vitro, respectively (88).
Although many studies have observed the aggravating or alleviating effects of circRNA on AMI in animal experiments. However, we need more experiment to knock out or overexpress specific circRNA in patients with AMI, and to observe its effect in human body for a long term, so as to explore targeted drugs for the treatment of AMI.
Involved in the differentiation of stem cell after AMI
Stem cell therapy has been widely used in regenerative medicine. A study has shown that during the differentiation of various types of stem cells, a large number of circRNAs, such as CDR1as, CircFOXP1 and CircZNF91, are dysregulated, which regulate the differentiation of different kinds of stem cells through the circRNA-miRNA-mRNA interaction network and various signaling pathways (such as PI3K Akt, MAPK and Wnt signal pathway) (98). Studies on cardiac regeneration have found that circRNA is involved in the proliferation and differentiation of cardiac cells, and participates in regulating the process of the differentiation of stem cells into cardiomyocyte-like cells. Most studies have focused on the regulation of circRNA during the process of differentiation of mesenchymal stem cells (MSCs) and induced pluripotent stem cells (iPSCs) (99,100). Lei et al. found that circRNA is highly enriched in cardiomyocytes derived from human iPS [a type of pluripotent stem cell (PSC)], so cardiac specific circRNA, including CircALPK2, CircCACNA1D, CircSLC8A1 and CircSPHKAP, can be used as biomarkers of cardiomyocytes (101). A study by Ruan et al. found that circRNA was differentially expressed in human umbilical cord-derived mesenchymal stem cells (hucMSCs) (102). Yang et al. found that the expression level of CDR1as was very high in hucMSCs, and knockdown of CDR1as inhibited the proliferation and differentiation potential of hucMSCs (103). Cherubini et al. showed that CircFOXP1, as a sponge of miR-17-3p and miR-127-5p, promoted the proliferation and differentiation of MSCs and was involved in regulating the differentiation of MSCs (99). In addition, CircRNA_05432, CircRNA_08441 and CircRNA_01536 were identified as key regulator during the differentiation of hucMSCs into cardiomyocyte-like cells (102). After AMI, due to the lack of clinically significant regeneration ability of mature cardiomyocytes, necrotic myocardial are usually replaced by non-functional scar tissue, but that’s not the best way to repair. Therefore, future research should focus on circRNA and the proliferation and differentiation of cardiomyocytes.
CircRNA as biomarker of AMI
At present, the main diagnostic markers for AMI are CK-MB and cTnT/I, but many diseases other than heart such as pulmonary embolism (PE) may interfere with the diagnosis (104). Therefore, we need to further explore biomarkers with higher sensitivity and specificity. CircRNAs have been proven to be effective biomarkers for many diseases, such as cancer, acquired pneumonia, and Kawasaki disease (105-107). The expression of circRNAs is supernal in patients with AMI and they may be sensitive biomarkers of AMI due to the stability and conservation (49). Therefore, highly stable, conserved and specific circRNAs may open up a new landscape for the diagnosis and prognosis of AMI (as summarized in Table 3).
According to bioinformatics analysis, the up-regulated CircTMEM165, CircUBAC2, CircZNF609, CircANKRD12 and CircSLC8A1 were highly related to MI, and these five circRNAs could be used to improve the diagnostic accuracy of MI (18). A study has confirmed that CircRNA_081881 in plasma could be used as a potential target for precise diagnosis and treatment of AMI (89). Zhao et al. showed that Hsa_circRNA_001654, hsa_circRNA_091761, hsa_circRNA_405624 and hsa_circRNA_406698 were significantly increased in the blood of patients with AMI, which could be used as potential biomarker for early diagnosis of AMI (90). The knockdown of CircROBO2 could reduce cardiomyocyte apoptosis by regulate miR-1184/TRADD axis, which may be a potential pathogenic gene of AMI and may provide evidence for the clinical prognosis of AMI (71). The different expression patterns of Circ-RCAN2 in the heart tissue of MI pigs suggest that Circ-RCAN2 has diagnostic significance for MI (91). The circRNA has_circ_0124644 was confirmed to be a potential biomarker. When has_circ_0124644 was introduced, the specificity and sensitivity of diagnosis were significantly improved for CVD (94). A meta-analysis identified CircCDKN2BAS and CircMACF1 were valuable biomarkers for CVD (108). But the last two studies did not specifically target AMI.
CircRNA as a biomarker has also been studied in forensic medicine. In acute ischemic heart disease (IHD) rat model, CircSLC8A1 was positively correlated with CK-MB in pericardial fluid and CircNFIX was negatively correlated with the degree of coronary artery stenosis. CircSLC8A1 and CircNFIX could be used as auxiliary diagnostic markers for SCD caused by IHD in forensic medicine (87). In terms of the prognosis of AMI, Vausort et al. found that patients with low expression of MICRA had a higher risk of LV dysfunction, and that MICRA could predict LV dysfunction three-four months after AMI and could be used for risk classification of AMI and early judgment of cardiac prognosis (92,93).
In general, with the in-depth study of circRNA, more and more circRNAs will be expected to become new biomarker, which will provide a more perfect strategy for the accurate diagnosis of AMI, medical identification in forensic medicine, risk classification and prognosis judgment after AMI. However, further studies are needed to compare the advantage of circRNA and cTn T/I, CK-MB, and NT-proBNP, and to confirm their clinical application.
Discussion and summary
With the further exploration of circRNA, new diagnostic and therapeutic method for AMI have attracted wide attention. Most evidence shows that circRNA is involved in and regulates the pathogenesis and pathophysiological of AMI mainly by regulating the circRNA/miRNA/mRNA axis. Silence or overexpress certain specific circRNAs can promote the process of cardiomyocyte apoptosis, fibrosis and other aggravating AMI, as well as the process of alleviating AMI such as angiogenesis and cardiac repair, which is a potential therapeutic target of AMI. Some specific circRNAs have also been identified as biomarker of AMI. In this paper, we systematically reviewed the biological characteristics of circRNA, involved in pathophysiology and cardiac repair of AMI, and is considered as a biomarker of AMI.
This review also has limitations. For example, we should notice that many studies on circRNA and AMI are still in the preliminary stage and focus on animal experiments, with mice or rats as specimens. Due to the complexity and expense of sequencing technology for circRNA, the sample size of sequencing is relatively small. However, the detection technology of cardiac troponin has been very mature and cheap. Therefore, we urgently need to improve the detection technology of circRNA, and more experiments in vivo are needed to observe the long-term influence of overexpress or silence some certain circRNA on cardiac function and prognosis after AMI, and further explore the clinical application value of genetic inhibitors or agonists for cardiac recovery after AMI.
It is also worth noting that circRNA and myocardial senescence after AMI may be a new field. In conclusion, circRNA has broad application prospects as specific biomarker and therapeutic target for AMI, which can be considered as an emerging role in AMI. We believe that circRNA will be widely used as a novel strategy for the diagnosis and treatment of AMI, and greatly reduce the adverse consequences of AMI.
Conclusions
CircRNA is mainly involved in and regulates the pathophysiology process of MI through different circRNA/miRNA/mRNA axes. Overexpressing or knockout certain specific circRNAs may aggravate or alleviate AMI. Some circRNAs have also been identified as biomarker of AMI. It has broad application prospects as specific biomarker and therapeutic targets for AMI, which can be considered as an emerging role in AMI.
Acknowledgments
Funding: This work was supported by the Medical Reserve Talents Funding Grant of Yunnan Health Commission (H-2018033).
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://jxym.amegroups.com/article/view/10.21037/jxym-23-10/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jxym.amegroups.com/article/view/10.21037/jxym-23-10/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dehghan M, Mente A, Zhang X, et al. Associations of fats and carbohydrate intake with cardiovascular disease and mortality in 18 countries from five continents (PURE): a prospective cohort study. Lancet 2017;390:2050-62. [Crossref] [PubMed]
- Yin L, Tang Y, Jiang M. Research on the circular RNA bioinformatics in patients with acute myocardial infarction. J Clin Lab Anal 2021;35:e23621. [Crossref] [PubMed]
- Anderson JL, Morrow DA. Acute Myocardial Infarction. N Engl J Med 2017;376:2053-64. [Crossref] [PubMed]
- Townsend N, Wilson L, Bhatnagar P, et al. Cardiovascular disease in Europe: epidemiological update 2016. Eur Heart J 2016;37:3232-45. [Crossref] [PubMed]
- Gulati R, Behfar A, Narula J, et al. Acute Myocardial Infarction in Young Individuals. Mayo Clin Proc 2020;95:136-56. [Crossref] [PubMed]
- Benjamin EJ, Virani SS, Callaway CW, et al. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 2018;137:e67-e492. [Crossref] [PubMed]
- Bugiardini R. Coronary Microcirculation and Ischemic Heart Disease, Today. Curr Pharm Des 2018;24:2891-2. [Crossref] [PubMed]
- Wu D, Zhang K, Hu P. The Role of Autophagy in Acute Myocardial Infarction. Front Pharmacol 2019;10:551. [Crossref] [PubMed]
- Gabriel-Costa D. The pathophysiology of myocardial infarction-induced heart failure. Pathophysiology 2018;25:277-84. [Crossref] [PubMed]
- Bougouin W, Marijon E, Puymirat E, et al. Incidence of sudden cardiac death after ventricular fibrillation complicating acute myocardial infarction: a 5-year cause-of-death analysis of the FAST-MI 2005 registry. Eur Heart J 2014;35:116-22. [Crossref] [PubMed]
- Gyöngyösi M, Winkler J, Ramos I, et al. Myocardial fibrosis: biomedical research from bench to bedside. Eur J Heart Fail 2017;19:177-91. [Crossref] [PubMed]
- Lu L, Liu M, Sun R, et al. Myocardial Infarction: Symptoms and Treatments. Cell Biochem Biophys 2015;72:865-7. [Crossref] [PubMed]
- Heallen T, Zhang M, Wang J, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 2011;332:458-61. [Crossref] [PubMed]
- Zhou Q, Li L, Zhao B, et al. The hippo pathway in heart development, regeneration, and diseases. Circ Res 2015;116:1431-47. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [Crossref] [PubMed]
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 2017;242:1136-41. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Li Q, Wang Y, An Y, et al. The Particular Expression Profiles of Circular RNA in Peripheral Blood of Myocardial Infarction Patients by RNA Sequencing. Front Cardiovasc Med 2022;9:810257. [Crossref] [PubMed]
- Cai H, Li Y, Niringiyumukiza JD, et al. Circular RNA involvement in aging: An emerging player with great potential. Mech Ageing Dev 2019;178:16-24. [Crossref] [PubMed]
- Ren S, Lin P, Wang J, et al. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol Ther Methods Clin Dev 2020;18:215-29. [Crossref] [PubMed]
- Cai L, Qi B, Wu X, et al. Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J Mol Cell Cardiol 2019;130:10-22. [Crossref] [PubMed]
- Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016;37:2602-11. [Crossref] [PubMed]
- Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 2016;7:12429. [Crossref] [PubMed]
- Altesha MA, Ni T, Khan A, et al. Circular RNA in cardiovascular disease. J Cell Physiol 2019;234:5588-600. [Crossref] [PubMed]
- Mei X, Chen SY. Circular RNAs in cardiovascular diseases. Pharmacol Ther 2022;232:107991. [Crossref] [PubMed]
- Wang L, Meng X, Li G, et al. Circular RNAs in Cardiovascular Diseases. Adv Exp Med Biol 2018;1087:191-204. [Crossref] [PubMed]
- Wu J, Li J, Liu H, et al. Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal 2019;33:e22998. [Crossref] [PubMed]
- Ding S, Zhu Y, Liang Y, et al. Circular RNAs in Vascular Functions and Diseases. Adv Exp Med Biol 2018;1087:287-97. [Crossref] [PubMed]
- Shan K, Liu C, Liu BH, et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017;136:1629-42. [Crossref] [PubMed]
- Miao R, Wang Y, Wan J, et al. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine (Baltimore) 2017;96:e7354. [Crossref] [PubMed]
- Zhou Y, Li C, Wang Z, et al. CircRNAs as Novel Biomarkers and Therapeutic Targets in Renal Cell Carcinoma. Front Mol Biosci 2022;9:833079. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015;160:1125-34. [Crossref] [PubMed]
- Liu C, Li N, Dai G, et al. A narrative review of circular RNAs as potential biomarkers and therapeutic targets for cardiovascular diseases. Ann Transl Med 2021;9:578. [Crossref] [PubMed]
- Xu T, Wu J, Han P, et al. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics 2017;18:680. [Crossref] [PubMed]
- Cortés-López M, Gruner MR, Cooper DA, et al. Global accumulation of circRNAs during aging in Caenorhabditis elegans. BMC Genomics 2018;19:8. [Crossref] [PubMed]
- Fischer JW, Leung AK. CircRNAs: a regulator of cellular stress. Crit Rev Biochem Mol Biol 2017;52:220-33. [Crossref] [PubMed]
- Zhang Z, Yang T, Xiao J. Circular RNAs: Promising Biomarkers for Human Diseases. EBioMedicine 2018;34:267-74. [Crossref] [PubMed]
- Tan WL, Lim BT, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res 2017;113:298-309. [Crossref] [PubMed]
- Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. [Crossref] [PubMed]
- Wilusz JE A. 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA 2018;9:e1478. [Crossref] [PubMed]
- Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792-806. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Corrigendum: Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2017;24:194. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [Crossref] [PubMed]
- Tian M, Cao Z, Pang H. Circular RNAs in Sudden Cardiac Death Related Diseases: Novel Biomarker for Clinical and Forensic Diagnosis. Molecules 2021;26:1155. [Crossref] [PubMed]
- Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol 2017;14:992-9. [Crossref] [PubMed]
- Tatomer DC, Wilusz JE. An Unchartered Journey for Ribosomes: Circumnavigating Circular RNAs to Produce Proteins. Mol Cell 2017;66:1-2. [Crossref] [PubMed]
- Schneider T, Bindereif A. Circular RNAs: Coding or noncoding? Cell Res 2017;27:724-5. [Crossref] [PubMed]
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 2017;27:626-41. [Crossref] [PubMed]
- Peng L, Chen G, Zhu Z, et al. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung's disease. Oncotarget 2017;8:808-18. [Crossref] [PubMed]
- Wen ZJ, Xin H, Wang YC, et al. Emerging roles of circRNAs in the pathological process of myocardial infarction. Mol Ther Nucleic Acids 2021;26:828-48. [Crossref] [PubMed]
- Aufiero S, Reckman YJ, Pinto YM, et al. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol 2019;16:503-14. [Crossref] [PubMed]
- Zhang S, Wang W, Wu X, et al. Regulatory Roles of Circular RNAs in Coronary Artery Disease. Mol Ther Nucleic Acids 2020;21:172-9. [Crossref] [PubMed]
- Sun JY, Shi Y, Cai XY, et al. Potential diagnostic and therapeutic value of circular RNAs in cardiovascular diseases. Cell Signal 2020;71:109604. [Crossref] [PubMed]
- Zhang L, Zhang Y, Yu F, et al. The circRNA-miRNA/RBP regulatory network in myocardial infarction. Front Pharmacol 2022;13:941123. [Crossref] [PubMed]
- Geng HH, Li R, Su YM, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PLoS One 2016;11:e0151753. [Crossref] [PubMed]
- Wang K, Gan TY, Li N, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 2017;24:1111-20. [Crossref] [PubMed]
- Wang S, Li L, Deng W, et al. CircRNA MFACR Is Upregulated in Myocardial Infarction and Downregulates miR-125b to Promote Cardiomyocyte Apoptosis Induced by Hypoxia. J Cardiovasc Pharmacol 2021;78:802-8. [Crossref] [PubMed]
- Liu X, Wang M, Li Q, et al. CircRNA ACAP2 induces myocardial apoptosis after myocardial infarction by sponging miR-29. Minerva Med 2022;113:128-34. [Crossref] [PubMed]
- Zhang J, Tang Y, Zhang J, et al. CircRNA ACAP2 Is Overexpressed in Myocardial Infarction and Promotes the Maturation of miR-532 to Induce the Apoptosis of Cardiomyocyte. J Cardiovasc Pharmacol 2021;78:247-52. [Crossref] [PubMed]
- Ye X, Hang Y, Lu Y, et al. CircRNA circ-NNT mediates myocardial ischemia/reperfusion injury through activating pyroptosis by sponging miR-33a-5p and regulating USP46 expression. Cell Death Discov 2021;7:370. [Crossref] [PubMed]
- Gan J, Yuan J, Liu Y, et al. Circular RNA_101237 mediates anoxia/reoxygenation injury by targeting let-7a-5p/IGF2BP3 in cardiomyocytes. Int J Mol Med 2020;45:451-60. [PubMed]
- Ni H, Li W, Zhuge Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol 2019;292:188-96. [Crossref] [PubMed]
- Zhai C, Qian G, Wu H, et al. Knockdown of circ_0060745 alleviates acute myocardial infarction by suppressing NF-κB activation. J Cell Mol Med 2020;24:12401-10. [Crossref] [PubMed]
- Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2017;38:1402-12. [PubMed]
- Li F, Long TY, Bi SS, et al. circPAN3 exerts a profibrotic role via sponging miR-221 through FoxO3/ATG7-activated autophagy in a rat model of myocardial infarction. Life Sci 2020;257:118015. [Crossref] [PubMed]
- Chen TP, Zhang NJ, Wang HJ, et al. Knockdown of circROBO2 attenuates acute myocardial infarction through regulating the miR-1184/TRADD axis. Mol Med 2021;27:21. [Crossref] [PubMed]
- Gao WQ, Hu XM, Zhang Q, et al. Downregulation of circFASTKD1 ameliorates myocardial infarction by promoting angiogenesis. Aging (Albany NY) 2020;13:3588-604. [Crossref] [PubMed]
- Liu H, Ma X, Mao Z, et al. Circular RNA has_circ_0003204 inhibits oxLDL-induced vascular endothelial cell proliferation and angiogenesis. Cell Signal 2020;70:109595. [Crossref] [PubMed]
- Li M, Ding W, Tariq MA, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 2018;8:5855-69. [Crossref] [PubMed]
- Zhou LY, Zhai M, Huang Y, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ 2019;26:1299-315. [Crossref] [PubMed]
- Ren Q, Li H, Wang X. The circular RNA ZNF292 alleviates OGD-induced injury in H9c2 cells via targeting BNIP3. Cell Cycle 2019;18:3365-77. [Crossref] [PubMed]
- Zhu Y, Pan W, Yang T, et al. Upregulation of Circular RNA CircNFIB Attenuates Cardiac Fibrosis by Sponging miR-433. Front Genet 2019;10:564. [Crossref] [PubMed]
- Bai M, Pan CL, Jiang GX, et al. CircRNA 010567 improves myocardial infarction rats through inhibiting TGF-β1. Eur Rev Med Pharmacol Sci 2020;24:369-75. [PubMed]
- Sun LY, Zhao JC, Ge XM, et al. Circ_LAS1L regulates cardiac fibroblast activation, growth, and migration through miR-125b/SFRP5 pathway. Cell Biochem Funct 2020;38:443-50. [Crossref] [PubMed]
- Zeng Y, Du WW, Wu Y, et al. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics 2017;7:3842-55. [Crossref] [PubMed]
- Zhang Q, Sun W, Han J, et al. The circular RNA hsa_circ_0007623 acts as a sponge of microRNA-297 and promotes cardiac repair. Biochem Biophys Res Commun 2020;523:993-1000. [Crossref] [PubMed]
- Zhu Y, Zhao P, Sun L, et al. Overexpression of circRNA SNRK targets miR-103-3p to reduce apoptosis and promote cardiac repair through GSK3β/β-catenin pathway in rats with myocardial infarction. Cell Death Discov 2021;7:84. [Crossref] [PubMed]
- Garikipati VNS, Verma SK, Cheng Z, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun 2019;10:4317. [Crossref] [PubMed]
- Zhao B, Li G, Peng J, et al. CircMACF1 Attenuates Acute Myocardial Infarction Through miR-500b-5p-EMP1 Axis. J Cardiovasc Transl Res 2021;14:161-72. [Crossref] [PubMed]
- Cheng N, Wang MY, Wu YB, et al. Circular RNA POSTN Promotes Myocardial Infarction-Induced Myocardial Injury and Cardiac Remodeling by Regulating miR-96-5p/BNIP3 Axis. Front Cell Dev Biol 2020;8:618574. [Crossref] [PubMed]
- Huang S, Li X, Zheng H, et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation 2019;139:2857-76. [Crossref] [PubMed]
- Tian M, Xue J, Dai C, et al. CircSLC8A1 and circNFIX can be used as auxiliary diagnostic markers for sudden cardiac death caused by acute ischemic heart disease. Sci Rep 2021;11:4695. [Crossref] [PubMed]
- Zhang M, Wang Z, Cheng Q, et al. Circular RNA (circRNA) CDYL Induces Myocardial Regeneration by ceRNA After Myocardial Infarction. Med Sci Monit 2020;26:e923188. [Crossref] [PubMed]
- Deng YY, Zhang W, She J, et al. GW27-e1167 Circular RNA Related to PPARγ Function as ceRNA of microRNA in Human Acute Myocardial Infarction. J Am Coll Cardiol 2016;68:C51-2. [Crossref]
- Zhao C, Liu J, Ge W, et al. Identification of Regulatory circRNAs Involved in the Pathogenesis of Acute Myocardial Infarction. Front Genet 2020;11:626492. [Crossref] [PubMed]
- Mester-Tonczar J, Einzinger P, Winkler J, et al. Novel Identified Circular Transcript of RCAN2, circ-RCAN2, Shows Deviated Expression Pattern in Pig Reperfused Infarcted Myocardium and Hypoxic Porcine Cardiac Progenitor Cells In Vitro. Int J Mol Sci 2021;22:1390. [Crossref] [PubMed]
- Vausort M, Salgado-Somoza A, Zhang L, et al. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J Am Coll Cardiol 2016;68:1247-8. [Crossref] [PubMed]
- Salgado-Somoza A, Zhang L, Vausort M, et al. The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc 2017;17:33-6. [Crossref] [PubMed]
- Zhao Z, Li X, Gao C, et al. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 2017;7:39918. [Crossref] [PubMed]
- Boeckel JN, Jaé N, Heumüller AW, et al. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res 2015;117:884-90. [Crossref] [PubMed]
- Wang L, Yu P, Wang J, et al. Downregulation of circ-ZNF609 Promotes Heart Repair by Modulating RNA N(6)-Methyladenosine-Modified Yap Expression. Research (Wash D C) 2022;2022:9825916. [Crossref] [PubMed]
- Mester-Tonczar J, Winkler J, Einzinger P, et al. Association between Circular RNA CDR1as and Post-Infarction Cardiac Function in Pig Ischemic Heart Failure: Influence of the Anti-Fibrotic Natural Compounds Bufalin and Lycorine. Biomolecules 2020;10:1180. [Crossref] [PubMed]
- Lin Z, Tang X, Wan J, et al. Functions and mechanisms of circular RNAs in regulating stem cell differentiation. RNA Biol 2021;18:2136-49. [Crossref] [PubMed]
- Cherubini A, Barilani M, Rossi RL, et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res 2019;47:5325-40. [Crossref] [PubMed]
- Mester-Tonczar J, Hašimbegović E, Spannbauer A, et al. Circular RNAs in Cardiac Regeneration: Cardiac Cell Proliferation, Differentiation, Survival, and Reprogramming. Front Physiol 2020;11:580465. [Crossref] [PubMed]
- Lei W, Feng T, Fang X, et al. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther 2018;9:56. [Crossref] [PubMed]
- Ruan ZB, Chen GC, Zhang R, et al. Circular RNA expression profiles during the differentiation of human umbilical cord-derived mesenchymal stem cells into cardiomyocyte-like cells. J Cell Physiol 2019;234:16412-23. [Crossref] [PubMed]
- Yang L, Bin Z, Hui S, et al. The Role of CDR1as in Proliferation and Differentiation of Human Umbilical Cord-Derived Mesenchymal Stem Cells. Stem Cells Int 2019;2019:2316834. [Crossref] [PubMed]
- Zhou Q, Zhang Z, Bei Y, et al. Circular RNAs as Novel Biomarkers for Cardiovascular Diseases. Adv Exp Med Biol 2018;1087:159-70. [Crossref] [PubMed]
- Li X, Wang Y, Han C, et al. Colorectal cancer progression is associated with accumulation of Th17 lymphocytes in tumor tissues and increased serum levels of interleukin-6. Tohoku J Exp Med 2014;233:175-82. [Crossref] [PubMed]
- Zhao T, Zheng Y, Hao D, et al. Blood circRNAs as biomarkers for the diagnosis of community-acquired pneumonia. J Cell Biochem 2019;120:16483-94. [Crossref] [PubMed]
- Wu J, Zhou Q, Niu Y, et al. Aberrant expression of serum circANRIL and hsa_circ_0123996 in children with Kawasaki disease. J Clin Lab Anal 2019;33:e22874. [Crossref] [PubMed]
- Li JJ, Wang W, Wang XQ, et al. A novel strategy of identifying circRNA biomarkers in cardiovascular disease by meta-analysis. J Cell Physiol 2019;234:21601-12. [Crossref] [PubMed]
Cite this article as: Zhao W, Zhu G, Chu T, Wu L, Li H. CircRNA as an emerging role in acute myocardial infarction: a literature review. J Xiangya Med 2023;8:9.